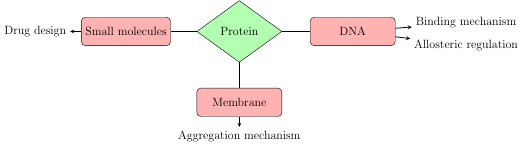
Use theoretical and molecular modeling approaches to understand:
1. Dynamic processes and biophysical basis of protein folding and aggregation.
2. Mechanisms of protein-DNA recognition.
3. Structure-based drug design.
4. Allosteric communication in protein-DNA complex.



